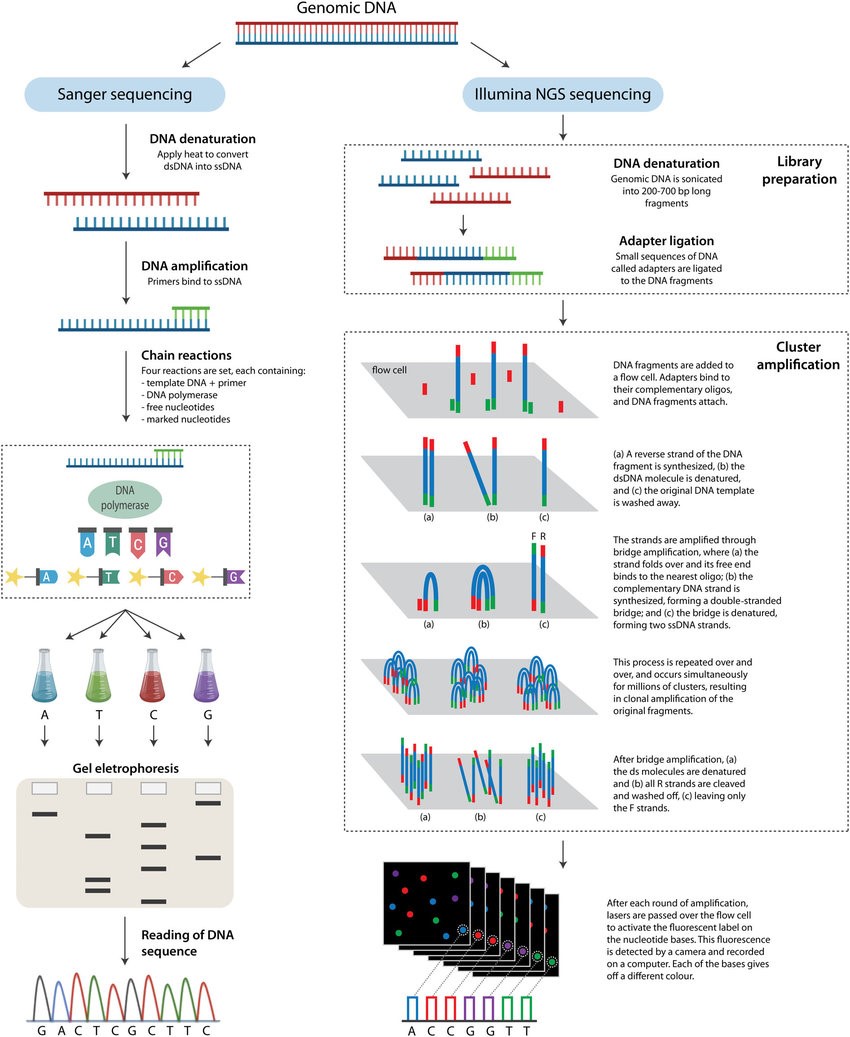
Next-generation sequencing (NGS) is a massively parallel sequencing technology that offers ultra-high throughput, scalability, and speed. The technology is used to determine the order of nucleotides in entire genomes or targeted regions of DNA or RNA.
NGS technology has fundamentally changed the kinds of questions scientists can ask and answer. Innovative sample preparation and data analysis options enable a broad range of applications. For example, NGS allows labs to:
• Rapidly sequence whole genomes
• Deeply sequence target regions
• Utilize RNA sequencing (RNA-Seq) to discover novel RNA variants and splice sites, or quantify mRNAs for gene expression analysis
• Analyze epigenetic factors such as genome-wide DNA methylation and DNA-protein interactions
• Sequence cancer samples to study rare somatic variants, tumor subclones, and more
• Study the human microbiome
• Identify novel pathogens
NGS in Plant Research:
Non-model plants i.e., the species which have one or all of the characters such as long life cycle, difficulty to grow in the laboratory or poor fecundity, have been schemed out of sequencing projects earlier, due to high running cost of Sanger sequencing. Consequently, the information about their genomics and key biological processes are inadequate. However, the advent of fast and cost-effective next-generation sequencing (NGS) platforms in the recent past has enabled the unearthing of certain characteristic gene structures unique to these species. It has also aided in gaining insight about mechanisms underlying processes of gene expression and secondary metabolism as well as facilitated development of genomic resources for diversity characterization, evolutionary analysis and marker-assisted breeding even without prior availability of genomic sequence information.
Next-generation sequencing, in contrast, makes large-scale whole-genome sequencing (WGS) accessible and practical for the average researcher. It enables scientists to analyze the entire human genome in a single sequencing experiment, or sequence thousands to tens of thousands of genomes in one year.
The article has been written with the collaborative research of Mr. Ataur Rahman, Scientist and Mr. Aqief Afzal, Research Assistant of ASRBC, ACI Seed, ACI Ltd.