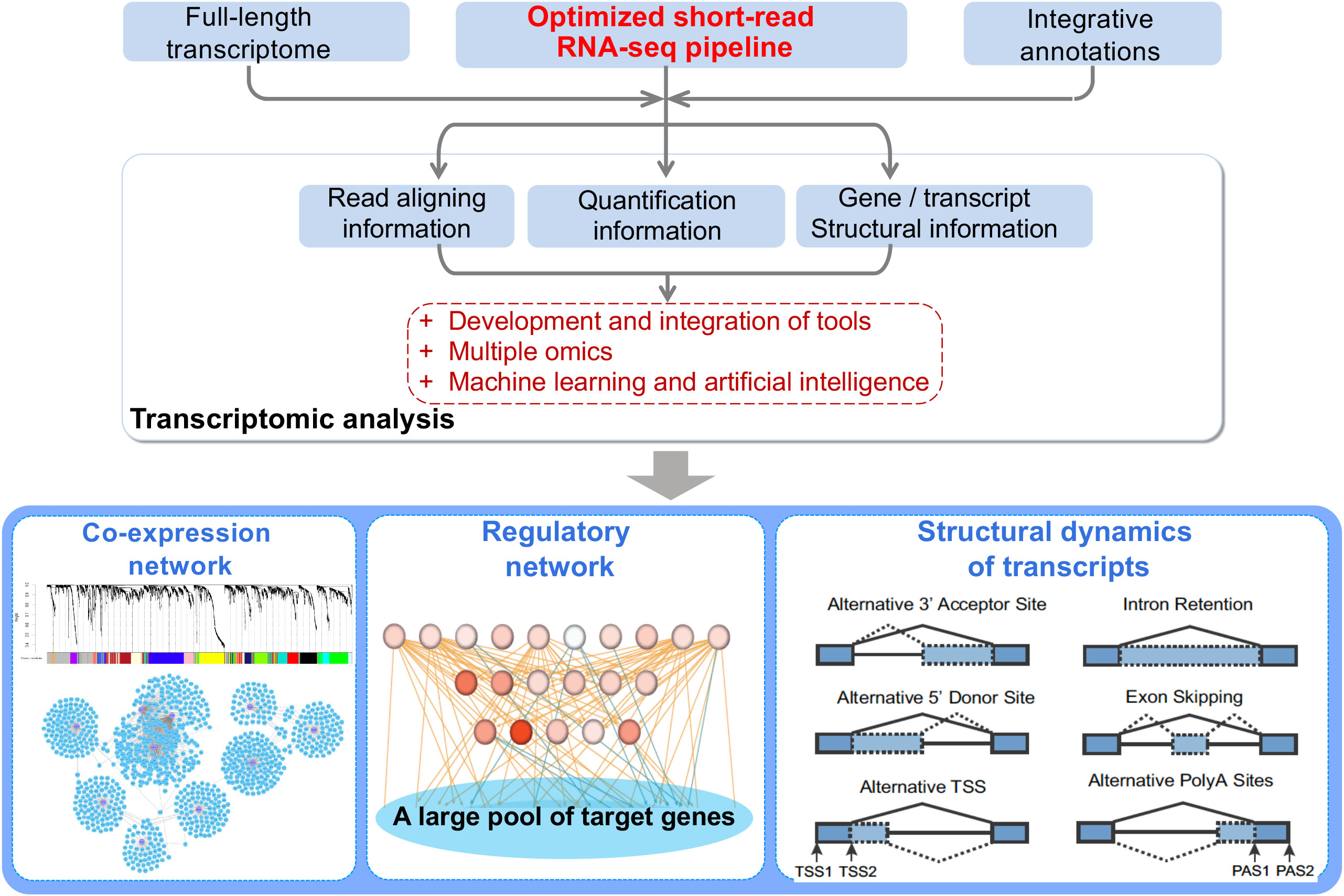
Transcriptomics plays a crucial role in the molecular breeding research of plants by providing valuable insights into the genetic and molecular mechanisms underlying desirable traits. Here's how transcriptomics is used in the context of plant molecular breeding:
- Trait Identification and Characterization: Transcriptomics helps identify candidate genes and regulatory elements associated with specific traits of interest, such as disease resistance, abiotic stress tolerance, enhanced nutritional content, or increased yield. By comparing gene expression profiles between plants with and without the desired traits, researchers can pinpoint key genes and pathways.
- Molecular Marker Development: Once candidate genes are identified through transcriptomics, researchers can develop molecular markers like single nucleotide polymorphisms (SNPs) or simple sequence repeats (SSRs) linked to these genes. These markers can be used for marker-assisted selection (MAS) in breeding programs to track and select plants with the target traits.
- Marker-Assisted Selection (MAS): Transcriptomics data can inform MAS by identifying differentially expressed genes associated with specific traits. Breeders can then use gene expression profiles and molecular markers to select plants with the desired traits more efficiently and accurately.
- Genomic Selection: By combining transcriptomics data with genomic data, such as DNA markers, researchers can improve the accuracy of genomic selection models. These models use genetic and gene expression information to predict the performance of potential breeding candidates, making breeding decisions more data-driven.
- Understanding Gene Regulation: Transcriptomics provides insights into the regulatory mechanisms that control gene expression. Understanding these mechanisms is crucial for fine-tuning gene expression and manipulating regulatory pathways to develop plants with specific traits.
- Quantitative Trait Loci (QTL) Analysis: Transcriptomics can aid in the identification of QTLs associated with important traits. By correlating gene expression data with specific traits, researchers can map QTLs more precisely and identify candidate genes for breeding.
- Stress Tolerance and Adaptation: Transcriptomics is instrumental in breeding crops with improved stress tolerance. It allows researchers to study the genetic responses of plants to various stressors, such as drought, salinity, pests, and diseases, leading to the development of stress-resistant varieties.
- Accelerated Breeding Cycles: Transcriptomics expedites the breeding process by enabling early selection of promising breeding candidates based on gene expression profiles. This accelerates breeding cycles and allows for the faster development of new crop varieties.
- Customized Breeding Programs: Transcriptomics enables the tailoring of breeding programs to specific environmental conditions and market demands. Breeders can select genes relevant to local conditions, resulting in plants better adapted to the target environment.
- Enhancing Nutritional Content: Transcriptomics can be used to identify and manipulate genes responsible for the production of essential nutrients, vitamins, and other bioactive compounds in crops. This is critical for developing plants with improved nutritional value.
- Environmental Sustainability: Transcriptomics can be employed to develop crops that are more resource-efficient, require fewer inputs (e.g., water, fertilizers, pesticides), and have lower environmental impacts.
In summary, transcriptomics is a powerful tool in plant molecular breeding research. It enables breeders to make informed decisions, accelerate the development of improved crop varieties, and address the challenges of food security, climate change, and sustainable agriculture by selecting and manipulating genes associated with desirable traits.
Dr. Md. Monirul Islam
Senior Scientist
ASRBC, ACI Seed