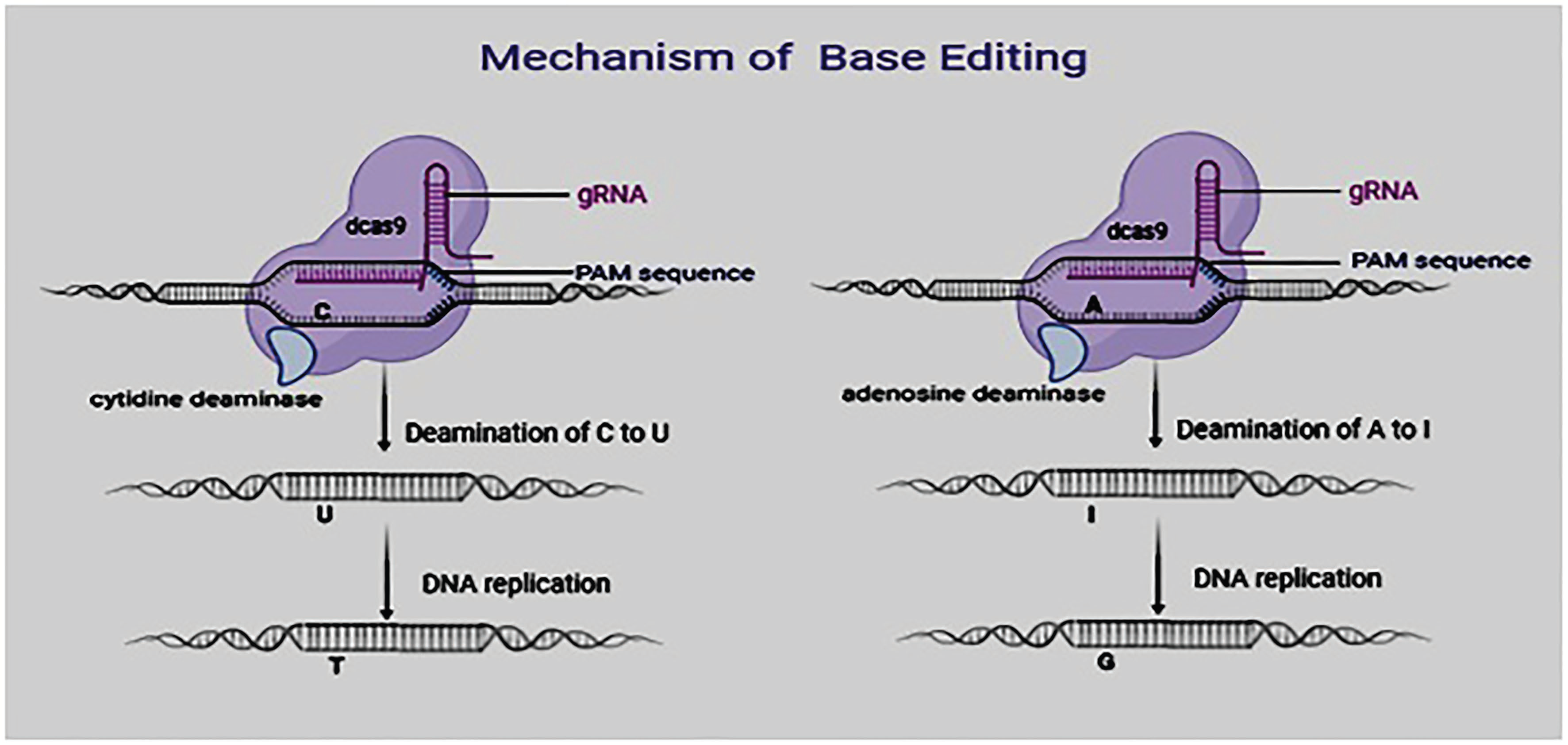
Base editing is a CRISPR-Cas9-based genome editing technique that allows point mutations in DNA to be introduced without causing double-strand breaks (DSBs). There are two types of base editors: cytidine base editors (CBEs) that enable C>T conversions and adenine base editors (ABEs) that allow A>G conversions. Base editors are larger than the standard CRISPR-Cas9 machinery, requiring more effort to synthesize mRNA and functional protein, and different AAV (Adeno-associated viruses) delivery strategies, such as using a two-part packaging system, or a smaller inactivated nuclease. This technology has attracted global attention and has been employed in functional analysis studies in crop plants (Azameti and Dauda 2021). Since many important agronomic traits are confirmed to be determined by single-nucleotide polymorphisms, improved crop varieties could be developed by the programmed and precise conversion of targeted single bases in the genomes of plants and animals. Because of its accuracy, simplicity, and multiplex capabilities, this technology has recently achieved rapid adoption and application. Unfortunately, the CRISPR/Cas9 system cannot be used to carry out gene base conversion. They are most appropriate in knock-out or knock-in of genes. Owing to these limitations, it is imperative to look for a precise and stable approach for editing crop/animal genomes. Base editing has been regarded as an alternative and more efficient approach (Veillet, Perrot et al. 2019). It overcomes some limitations of CRISPR/Cas9 by its utilization of a tethered deaminase domain or nickase Cas9 for base conversion from A > G or C > T. Recent studies have utilized this technique to create both single and multiple nucleotide modifications in cells (Yan, Ren et al. 2021). Base editing is the targeted substitution of one base or base pair with another that does not result in DSBs. This is done using either DNA or RNA base editors. A base editor consists of an inactive CRISPR–Cas9 component (Cas9 variants, dCas9, or Cas9 nickase) and a deaminase (cytosine or adenosine) component that converts one base to another.
Dr. Md. Monirul Islam
Senior Scientist
ASRBC, ACI Seed
References:
Azameti, M. K. and W. P. Dauda (2021). "Base editing in plants: applications, challenges, and future prospects." Frontiers in Plant Science: 1531.
Veillet, F., et al. (2019). "Transgene-free genome editing in tomato and potato plants using Agrobacterium-mediated delivery of a CRISPR/Cas9 cytidine base editor." International Journal of Molecular Sciences 20(2): 402.
Yan, D., et al. (2021). "High-efficiency and multiplex adenine base editing in plants using new TadA variants." Molecular Plant 14(5): 722-731.