ChIP-Seq data plays a crucial role in plant breeding by providing valuable insights into the regulatory mechanisms that govern gene expression and influence various traits. Here are several ways in which ChIP-Seq data is important for advancing plant breeding:
- Identification of Regulatory Elements: ChIP-Seq allows the identification of regulatory elements such as transcription factor binding sites and histone modifications associated with active or repressed chromatin regions. Understanding the regulatory landscape provides insights into how genes controlling important traits are regulated.
- Characterization of Transcription Factor Networks: ChIP-Seq helps in characterizing transcription factor networks by identifying the specific binding sites of transcription factors across the genome. This information is critical for unravelling the complex gene regulatory networks that influence traits relevant to plant breeding.
- Insights into Epigenetic Modifications: ChIP-Seq can profile epigenetic modifications such as DNA methylation and histone modifications. Epigenetic modifications play a role in heritable changes in gene expression and can impact traits important for plant breeding.
- Understanding Chromatin Dynamics during Development: ChIP-Seq enables the study of dynamic changes in chromatin structure during different developmental stages. Understanding how chromatin changes during development provides insights into the regulation of traits at different growth phases.
- Differential Analysis for Genotypic and Phenotypic Variation: ChIP-Seq data can be used for differential analysis between different genotypes or phenotypic states. Identifying differential binding patterns helps in understanding the molecular basis of phenotypic diversity and informs breeding strategies.
- Marker Discovery and Marker-Assisted Selection: ChIP-Seq data can contribute to the discovery of molecular markers associated with desirable traits. These markers can be used in marker-assisted selection (MAS) to accelerate the breeding process by selecting individuals with the desired genomic characteristics.
- Optimizing Breeding Strategies: ChIP-Seq data provides insights into the regulatory elements controlling key traits, enabling breeders to design more targeted and efficient breeding strategies. This information helps in selecting parents with the desired regulatory profiles to enhance the probability of obtaining desired traits in the offspring.
- Precision Breeding for Stress Tolerance: ChIP-Seq can be applied to study the regulatory mechanisms involved in stress response pathways. This information is crucial for developing crops with improved stress tolerance, a key goal in plant breeding to ensure productivity under changing environmental conditions.
- Integration with Other Omics Data: Integration of ChIP-Seq data with other omics data (e.g., RNA-Seq, metabolomics) provides a comprehensive understanding of the molecular processes underlying trait expression. ChIP-Seq data contributes significantly to our understanding of the molecular mechanisms controlling traits relevant to plant breeding. By deciphering the regulatory code embedded in the genome, breeders can make informed decisions, leading to more efficient and targeted crop improvement strategies.
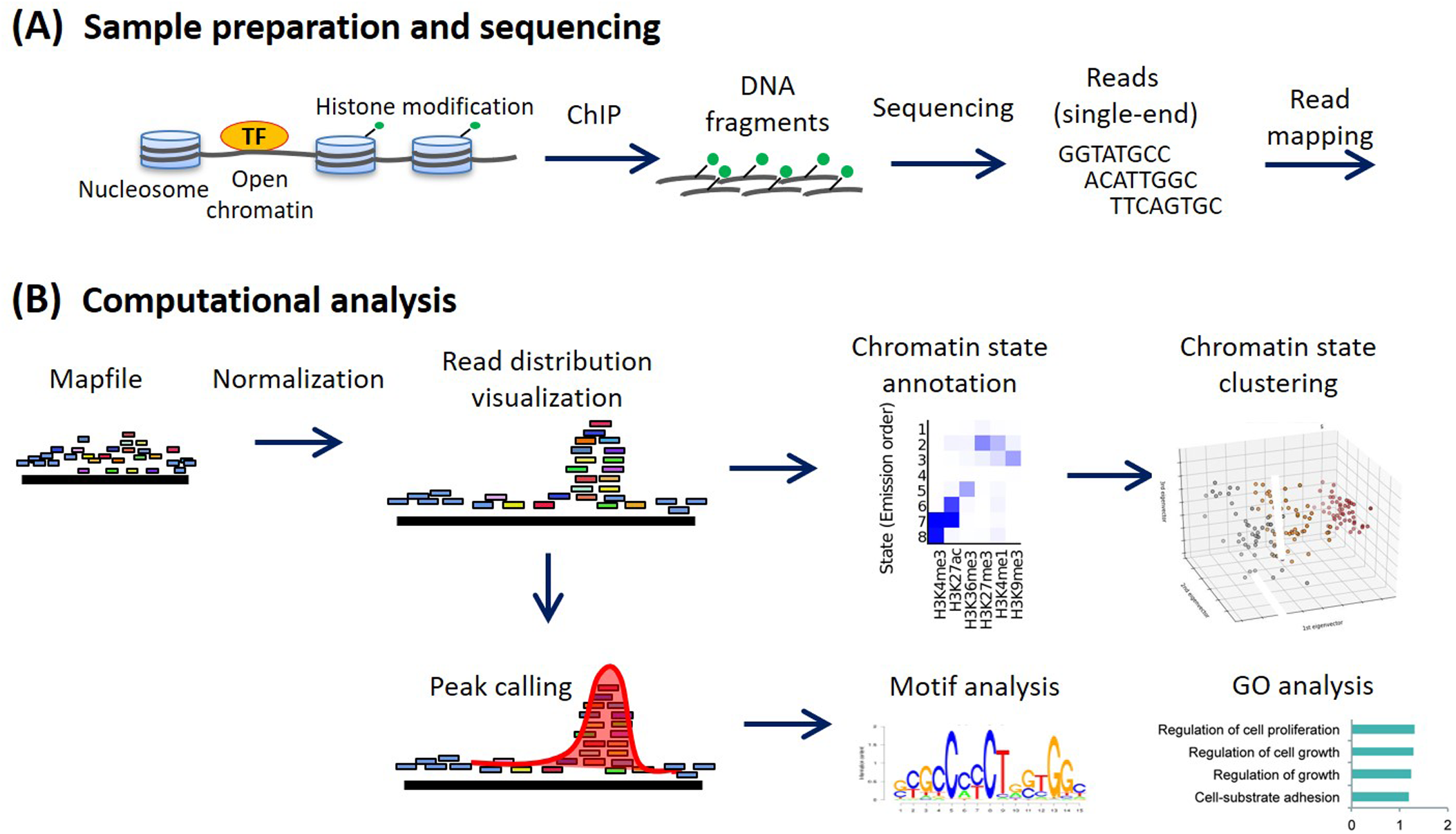
Fig. ChIP-seq analysis workflow. (A) Sample preparation and sequencing. (B) Computational analysis in a canonical ChIP-seq analysis. Various analyses are implemented using normalized read distribution.
Dr. Md. Monirul Islam
Senior Scientist
ASRBC, ACI Seed



