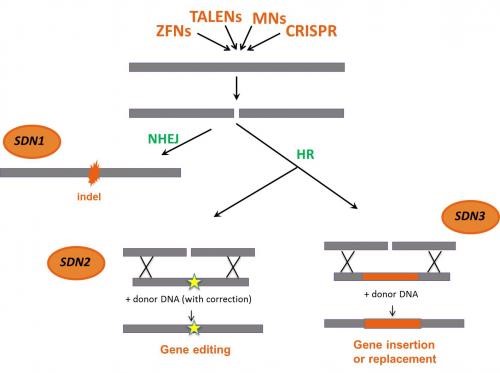
SDN is the collective term for the technique of creating a specific break in DNA with a nuclease, an enzyme that cuts DNA. Targeted modifications to the plant's traits are intended to be introduced by using the DNA break and the plant's inherent repairing systems.
Site-directed nucleases (SDNs) are a group of molecular tools used for targeted genome modification. They are designed to induce precise changes at specific locations within the genome of an organism. This technology has revolutionized genetic engineering and holds immense potential in various fields including biotechnology, medicine, and agriculture. There are several types of site-directed nucleases commonly used:
-
Zinc Finger Nucleases (ZFNs): ZFNs are engineered proteins composed of a DNA-binding domain derived from zinc finger proteins and a nuclease domain, typically from the FokI endonuclease. The DNA-binding domain is designed to recognize a specific DNA sequence, enabling precise targeting of desired genomic loci.
-
Transcription Activator-Like Effector Nucleases (TALENs): TALENs are another type of engineered nuclease composed of a DNA-binding domain derived from transcription activator-like effectors (TALEs) found in certain plant pathogenic bacteria. Similar to ZFNs, TALENs can be customized to recognize specific DNA sequences and induce double-strand breaks (DSBs) at desired genomic sites.
-
CRISPR-Cas System: Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and CRISPR-associated (Cas) proteins comprise an adaptive immune system found in bacteria and archaea. In recent years, the CRISPR-Cas system has been repurposed as a powerful tool for genome editing. The Cas nuclease, such as Cas9 or Cas12a, can be programmed with a guide RNA (gRNA) to target specific DNA sequences within the genome, resulting in precise genome modifications.
SDNs function by creating double-strand breaks (DSBs) at specific genomic locations. The cell's natural repair mechanisms then kick in to either repair the break via non-homologous end joining (NHEJ), often resulting in small insertions or deletions (indels) that disrupt the target gene's function, or through homology-directed repair (HDR), which can be used to introduce specific changes or insertions using a donor DNA template.
• In SDN-1 applications, mutations consisting of changes in a few base pairs, short deletions, or insertions (indels) are generated in a predefined region in the genome as a result of error-prone gene repair mechanisms of the cell (NHEJ). The repair mechanism does not require exogenous delivered DNA.
• In SDN-2 applications, specific point mutations, and small deletions/additions are generated as a result of the introduction into the cell of a repair DNA template (donor DNA) homologous to the targeted area. Using homologous recombination (HR), precise and small genetic modification can be achieved.
• In SDN-3 applications, entire genes can be inserted into a desired site in the genome. This is enabled by the delivery of a large stretch of recombinant DNA molecule (an exogenous donor DNA up to several kilobases long). The insertion can take place either by HR or by NHEJ.
At the molecular level, the genetic alterations produced by SDN-1 and SDN-2 are identical to those produced by irradiation, chemical mutagenesis, or spontaneous natural mutations and cannot be distinguished from them. The genetic alterations are brought about by the host's cellular DNA repair systems, which also fix breaks in DNA caused by natural processes. The CRISPR-Cas system, in particular, has gained widespread popularity due to its simplicity, versatility, and efficiency compared to ZFNs and TALENs. However, each system has its advantages and limitations, and the choice of which to use depends on factors such as the specific application, target organism, and desired outcomes.
Source: https://www.farm-europe.eu/travaux/new-plant-breeding-techniques-what-are-we-talking-about/